DNA methylation, knock out studies, site directed mutagenesis, transgenic mice, enzymology.
Abstract
DNA methylation is an essential process in mammals involved in several instances of gene regulation. So far, three active DNA methyltransferases have been found in mammals, Dnmt1 involved in maintenance methylation and Dnmt3a and 3b involved in de novo DNA methylation. Dnmt1 is a large protein comprising 1620 amino acid residues. Complete knock-out of the Dnmt1 enzyme is lethal during embryogenesis. Reduction of Dnmt1 causes demethylation of the DNA in vivo. It causes an arrest of DNA replication and and induces apoptosis. If these effects are due to demethylation is not known. Dnmt1 is phosphorylated and interacts with several other proteins in vitro. In vitro evidence suggests that it might also be implicated in a process called spreading of methylation. In this application we intend to prepare Dnmt1 variants carrying single or few amino acid exchanges that specifically address defined biochemical functions and characterize the mutant enzymes in vitro. The in vitro results will be compared with results from experiments with transgenic animal which contain the same defined exchanges in their endogenous dnmt1 gene. Such experiments so far have not been carried out with any DNA methyltransferase. Following this combined approach, we attempt to investigate a) if the loss of catalytic activity of Dnmt1 is responsible for embryonic lethality of the full knock-out, b) if the interaction of Dnmt1 with the PCNA protein has a role in vivo, c) if phosphorylation of Dnmt1 is of relevance in vivo and d) if an allosteric activation of Dnmt1 for methylation of unmodified DNA that has been observed in vitro and might cause spreading of methylation has a function in vivo. Finally, we attempt to investigate the biological role of an additional DNA methyltransferase, Dnmt2, whose function so far is not known by preparing a mouse knock-out strain and carefully investigate the transgenic animals over several generations. We expect that the results of these experiments will considerably further our knowledge on DNA methylation and epigenetic gene control.
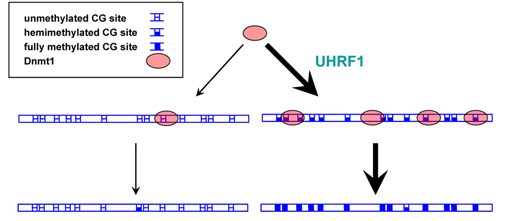 Double sieve mechanism to ensure accurate maintenance methylation based on Dnmt1 recruitment and specificity. UHRF1 preferentially recruits Dnmt1 to hemimethylated DNA regions, which had been methylated before DNA replication. Dnmt1 displays higher catalytic activity on hemimethylated DNA as compared to unmethylated DNA. (taken from Jeltsch, A. (2008) Reading and writing DNA methylation. Nat. Struct. Mol. Biol. 15, 1003-4).
Double sieve mechanism to ensure accurate maintenance methylation based on Dnmt1 recruitment and specificity. UHRF1 preferentially recruits Dnmt1 to hemimethylated DNA regions, which had been methylated before DNA replication. Dnmt1 displays higher catalytic activity on hemimethylated DNA as compared to unmethylated DNA. (taken from Jeltsch, A. (2008) Reading and writing DNA methylation. Nat. Struct. Mol. Biol. 15, 1003-4).
| Keywords: | Methylation, Knock out | |
| Domain: | Molecular Epigenetics | |
| Link: | http://www.jacobs-university.de/schools/ses/ajeltsch/ |
Publications
Zhang, Y., Rohde, C., Tierling, S., Jurkowski, T.P., Bock, C., Santacruz, D., Ragozin, S., Reinhardt, R., Groth, M., Walter, J., & Jeltsch, A. (2009) DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolutionm, PLOS Genetics, in press.
Jurkowska, R. Z., Anspach, N., Urbanke, C., Jia, D., Reinhardt, R., Nellen, W., Cheng, X. & Jeltsch, A. (2008) Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 36, 6656-63.
(…)
Jurkowska, R. Z., Anspach, N., Urbanke, C., Jia, D., Reinhardt, R., Nellen, W., Cheng, X. & Jeltsch, A. (2008) Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 36, 6656-63.
(…)
Contact
Prof. Dr. Albert
Jeltsch
Professor of Biochemistry
Research II
Jacobs University Bremen
Campus Ring 1
28759 Bremen, Germany
Phone: +49 421 200 3247
Fax: +49 421 200 3249
Email:
Professor of Biochemistry
Research II
Jacobs University Bremen
Campus Ring 1
28759 Bremen, Germany
Phone: +49 421 200 3247
Fax: +49 421 200 3249
Email: